I often tell undergraduates about the importance of conducting research projects in their senior year if they intend to pursue graduate studies in science. There are two main reasons for this. The first is that the labs that they have experienced up to that point in undergraduate courses, though useful for introducing specific concepts, are a poor reflection of what real science is like. As such, it is important for them to experience original lab work, rather than simply following a pre-defined, “cook book” protocol with an expected result. Novel studies have no pre-defined sequence, no a priori expectation of the outcome, and in many cases no established methods in place for generating data. The second is that there are some important lessons that they need to learn about research, perhaps most importantly that whatever they try to do in the lab will not work the first time. The sooner they hit that wall — and get around it — the better. Nature does not give up her secrets easily, I sometimes say.
My first experience with original research came during my fourth year at McMaster University (Hamilton, Canada), though the story actually begins the year before. In the late 1990s, McMaster offered a course on “environmental physiology” which explored the various ways that animals had become adapted to different extreme environments and lifestyles. For example, how insects can survive in deserts or how deep diving mammals conserve oxygen. I was very interested in organism biology and wanted to take this course, but it was a senior course and was only offered in alternate years. This meant taking it in my 3rd year without the prerequisites, which the instructors agreed to let me (and my roommate) do, as long as we took the prerequisities after the fact.
There were fewer than 10 people in the class (several of whom were graduate students) as it had a reputation for being very intense, with long labs on Friday evenings. Because of the small class size, we came to know the professors rather well, and I naturally asked them to supervise my undergraduate thesis project the following year. Specifically, I worked with Dr. Chris Wood, who is a very well regarded fish physiologist. Most of the work in his lab at the time had to so with metal toxicity, waste excretion, and so on, but one area struck me as particularly interesting — it turned out that an earlier study had suggested that fish who grew the most rapidly did not swim well. Being interested in evolution and organism biology, the notion of a trade-off between growth and swimming seemed like a very interesting issue to explore. I proposed a project that would look at this possible trade-off between growth and swimming, but also would include a component of feeding competition and social hierarchy (Who gets the most food? Who grows fastest? Do dominant fish swim worse than subordinate fish?).
There were, of course, numerous obstacles to overcome. How could I identify individual fish? How could I measure dominance rank and feeding? How should swimming performance be assessed? What size of fish should be used? And so on. After much trial and error, I settled on a system for identifying fish using a coded system of ink dots on the skin which were made using an injector that was once used to inject anaesthetic into the gums of dental patients. Lesson 1: Be prepared to be creative in terms of what counts as a scientific apparatus. (Dr. Wood once published a study conducted in Kenya which described the fish as being kept in “amber Tusker chambers” — Tusker being the local beer).
method I developed. Pretty decent for Windows Paint, if I may say so myself.
Growth rate was relatively straightforward in principle: weigh and measure the lengths of the fish at regular intervals. Fortunately, someone else in the lab had constructed a “fish measuring tube”, which was a transparent plastic half-cylinder with a ruler under it, on a slant with drain holes at the bottom. Worked a treat.
Swimming performance was assessed using a huge swim tunnel that had been built years before, but which was modified so I could reach in and get the fish to keep swimming once they had become stuck against the grating at the end of the swim section. I measured both maximum sustainable velocity (called Ucrit) as well as burst swimming.
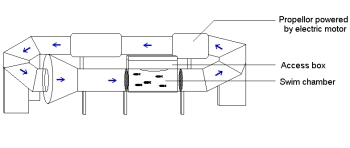
 Diagram from my thesis showing the swim tunnel apparatus.
Diagram from my thesis showing the swim tunnel apparatus.

The actual swim tunnel, which we knew as “Big Bertha”.
The quantification of feeding rates of individual fish (and by extension, their dominance rank) probably represented the most unusual component of the study. These were assessed by feeding the fish opaque glass beads mixed into the food and x-raying them so the beads in their stomachs could be counted. McMaster’s biology department is adjacent to a hospital, so I was able to secure the use of a small, portable x-ray machine that I could cart over to the lab on weekends. To make bead-laden food, I ground up fish pellets and mixed them with beads in a spaghetti maker, extruded the mush into strands and cut them into small pieces. I had to prepare all the food this way as a control, but I only gave them food with beads on days when they were to be x-rayed. To develop the x-ray images, I would go into the dark room in the radiology depatment of the hospital. This involved feeding the exposed films through an automatic developing machine which dropped the developed images into a slot outside the dark room. More than once I came out to find radiologists glancing curiously at my pictures of fish.

A typical x-ray film showing the difference in feeding among individual fish.

Close up of one fish showing the beads (white spots) in its stomach.
My first attempt at the study did not go well. The fish I had chosen were too small, and some even escaped from the swim chamber. I had chosen too high a ration to feed the fish, and the competition among individuals was negligible. I even had a flood in one tank and came in to find several fish flopping around on the floor before I returned them to their tank. Lesson 2: Be prepared for even the most carefully planned experiment to be a bust at first. As a matter of fact, I decided to start again after the holidays with larger fish and different rations. Given the short time involved (two semesters, one of which had then passed), it took some convincing for Dr. Wood to let me start from scratch. However, I knew I would not be confident in the data from the first run and I was fairly certain I had worked out the bugs the first time through.
In the end, we discovered that the fish who eat the most and grow the fastest do indeed show poorer sustained swimming performance, but only in the case where the ration is limited. Faster growing fish have better burst swimming abilities, by contrast. This supports the idea that there may be trade-offs between rapid growth and some types of swimming ability, at least when food is limited, meaning that there may be limits on the benefits of growing more quickly than other individuals.
I worked in the Wood Lab during the summer after this study and did two follow-up experiments. We also polished up my thesis and submitted it to a journal — I still remember how excited I was to have my first paper in review. Some major revisions later, the paper was accepted for publication. Lesson 3: Be prepared for peer reviewers to pick apart every detail of your work. (Not to worry, as I have since reviewed several fish physiology and feeding behaviour papers in turn).
I finished these studies more than a decade ago, and I now work in a very different area of biology. Nevertheless, the lessons that I learned during my first experience with original research are as significant as ever now that I advise students. If I have done my job well in this role, you may read a similar account from one of my students 10 years hence.
________
Gregory, T.R. and C.M. Wood. 1998. Individual variation and interrelationships between swimming performance, growth rate, and feeding in juvenile rainbow trout (Oncorhynchus mykiss). Canadian Journal of Fisheries and Aquatic Sciences 55: 1583-1590.
Gregory, T.R. and C.M. Wood. 1999. Interactions between individual feeding behaviour, growth, and swimming performance in juvenile rainbow trout (Oncorhynchus mykiss) fed different rations. Canadian Journal of Fisheries and Aquatic Sciences 56: 479-486.
Gregory, T.R. and C.M. Wood. 1999. The effects of chronic plasma cortisol elevation on the feeding behaviour, growth, competitive ability, and swimming performance of juvenile rainbow trout. Physiological and Biochemical Zoology 72: 286-295.










