The word logodaedaly means “a capricious coinage of words”. It was coined by Plato in the 4th century BC (as “wordsmith”) and picked up by Ben Johnson in 1611 in its current English usage. That’s right, someone coined a term for the process of coining terms.
Sometimes new terms are very useful. Every profession has its own jargon, which for the most part helps experts to save time by having individual terms for specific items or ideas. On the other hand, the original meaning can be lost and the term can be badly misunderstood or misapplied when it moves from jargon to buzzword. “Junk DNA” is a case in point. Other terms may be coined to give a simple summary of a more complex idea. “The Onion Test” is an example: it’s not really about onions, but about providing a reminder that there is more diversity out there than one might otherwise have considered.
Finally, sometimes terms are coined just for fun. This is one of those times.
Several bloggers have drawn attention to the persistent assumption expressed by some authors that humans are the pinnacle of biological complexity, as reflected in certain graphical representations relating to non-coding DNA [Pharyngula, Sandwalk, Sunclipse, Genomicron]. Larry Moran’s discussion pointed to what must be the single worst figure of the genre, from an article in Scientific American. This figure forms the basis of a new term that I wish to coin.
Here is the figure in question: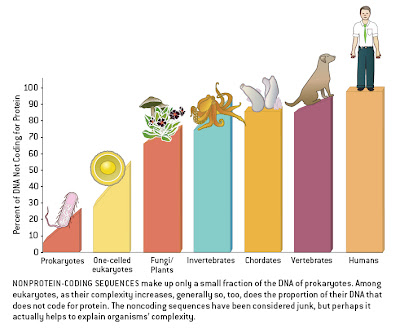
In a previous post, I complained about the ridiculous division of groups (humans are vertebrates and vertebrates are chordates), the lack of labels on the X-axis, the ambiguous definition of “complexity” implied, and the blatant assumption, sans justification, that humans are the most complex organisms around.
I also noted the following issue:
The sloping of the bars within taxa suggests that this is meant to imply a relationship between genome size and complexity within groups as well, with the largest genomes (i.e., the most non-coding DNA) found in the most complex organisms. This would negate the goal of placing humans at the extreme, as our genome is average for a mammal and at the lower end of the vertebrate spectrum (some salamanders have 20x more DNA than humans). Indeed, the human datum would accurately be placed roughly below the dog’s ass in this figure if it included a proper sampling of diversity.
As a result, I hereby propose that all such figures, with unlabeled axes and clear yet unjustified assumptions about complexity, henceforth be dubbed “Dog’s Ass Plots”. “DAPs” or “Dappers” also are acceptable, as in “I’m surprised that the reviewers didn’t pick up on this DAP” or “Check out this figure, it’s a real Dapper”. (As an added bonus, “dapper” means “neat and trim” — which these figures certainly are; the problem is not that they don’t look slick, it’s that they are oversimplified).
I have no doubt that plenty of examples can be found in subjects besides genomics, so please feel free to use it as needed in your own field.

I posted some ambiguous examples at Are These Dappers?. What do you think?
Ladies and gents, Larry Moran — the king of finding awful graphs!
Ok, I am making this post mandatory for my students – that is terrific.
(from a fellow prof down the road at U Waterloo)
Hmmm, you might to google the term DAP where you’ll probably find it has an established, quite different meaning. One I’m sure many Fundies are more familiar with especially given the recent troubles of the “Family Values” leaders.
Hmmm, you might to google the term DAP where you’ll probably find it has an established, quite different meaning. Can you clarify please? Download Accelerator Plus, DAP – Caulks, Sealants, Insulating Foam, Spackling, Glazing,…, Diagnostic Accreditation Program, Distributed Art Publishers, Digital audio player, Digital Archive Project….I hink dog’s ass plot is perfect.
Ooooh….can I talk about these in my grad seminar down the road at Western? Pretty please?
Sure, Katie. Just make sure you explain why they’re called DAPs!
Hey. Thank you for this info sharing. I guess this is a great ‘acronym’ or jargon name. I have not heard any of DAP before but it is worthwhile to use it, especially for those graphs without labeled axes etc. :D…paul
I disagree with some of the objections to this graph. It doesn’t matter that humans are vertebrates, or that vertebrates are chordates: the average for all vertebrates can be different from the average for humans. It doesn’t matter that the x-axis is not labeled if each of the bars are labeled. It makes sense to order the bars simply according to their height, to make it easier to find the highest, the lowest, and the middle-sized.
The caption goes on to draw the conclusion you object to, making the assumption that the categories represent increasing complexity to the right, but that assumption is not implicit in the figure. Couldn’t one use the figure to demonstrate that intelligence correlates with %DNA-that-does-not-code-for-protein? Or is it not as obvious as it seems to a lay person that average intelligence of those categories increases to the right.
One could measure car-size in different locations for environmental or marketing purposes, and make a table listing average size in regions such as North America, US, Canada, Toronto, Europe, UK, London, etc., and then rank the table according average size. There is nothing wrong with such a table, or bar graph generated from it. Even if the x-axis doesn’t correspond to any independent metric, the graph provides useful information about where the bigger cars are by continent, country, and city. And ranking the bars by height makes the information easier to extract. It also allows the audience to search for characteristics that might correlate with car size; characteristics that might have escaped the original publishers of the graph.
The entire range in genome size among eukaryotes is found among single-celled groups. Dogs are given as a representative of “vertebrates”, as distinct from humans, but the mammal range is relatively limited so they are not far from the human value. It absolutely matters that humans are somehow considered completely different from other vertebrates, presumably in terms of how complex they are — but there are lots of vertebrates with larger genomes. That fact destroys the relevance of the figure regardless of what your preferred comparison is. Your car analogy is not quite correct. It would be like plotting the following, with no axes, on the same graph:
European cars
Asian cars
North American cars
Ford cars
Ford Taurus
What sense would that make?
Far be it from me to argue about biology. I’m happy to accept your claim that for its stated purpose, this particular graph is ineffective, misleading, or ridiculous. But I understood your piece as an attempt to ridicule the graph based on the violation of what you consider to be general rules (or why coin a term?): (1) classifications in a bar graph must not be subsets of other classifications; (2) every x-axis must be identified with a quantifiable metric. My counter-example was entirely correct in that it violates both rules, but is a completely acceptable, useful, and common type of illustration.
If I’ve misrepresented your general rules, perhaps you can state them in a general way without reference to the particular graph, to see if they warrant coining a term.
And I don’t agree that in the offending graph, humans are “somehow considered completely different from other vertebrates, presumably in terms of how complex they areâ€, any more than the graph I proposed would imply London is somehow considered completely different from the UK, or in yours, that the Taurus is somehow considered completely different from North American cars. In fact, the graph you suggest might be an interesting comparison of car-size by different manufacturers by location and company. The inclusion of the Taurus would provide a convenient point of comparison, since we all have a pretty clear idea of its size. I’d guess the average Toyota is smaller than a Taurus, but this graph (if you included Toyota), would tell me by how much. Whether or not this information is useful to sociologists, environmentalists, or marketers, I don’t know, but that this type of information is usefully displayed in this type of illustration is not in any doubt, in my opinion.
If you implied that London was the endpoint of some undefined continuum without axes, then it would be a DAP.
My definitions have not been ambiguous.
In the current post:
“I hereby propose that all such figures, with unlabeled axes and clear yet unjustified assumptions about complexity, henceforth be dubbed “Dog’s Ass Plots”
And in a later one:
“Dog’s Ass Plot (DAP, or Dapper):
A graphical representation of data in any field that, through a lack of clear axis labels, selective inclusion/exclusion of data, visual presentation style, and/or other questionable characteristics, generates a misleading interpretation of the data in the viewer, especially by implying an illusory pattern that is not supported by the available data.”
“If you implied that London was the endpoint of some undefined continuum without axes, then it would be a DAP.â€
It wouldn’t be London, but some category would have the largest cars, and it would be plotted at the extreme right or left. Now without additional labels, is that implying that it is the endpoint of a continuum (other than that it has the largest cars)? Or is a DAP only a DAP if accompanied by a narrative supplying the offending implication?
“In the current post:

”I hereby propose that all such figures, with unlabeled axes and clear yet unjustified assumptions about complexity, henceforth be dubbed “Dog’s Ass Plots”

â€
I think you’re unclear on the concept of category bar graphs. If the categories are labeled, I’d argue they don’t need an additional x-axis label. And it’s not just me. I don’t know if there’s an authority on these matters, but my Penguin Dictionary of Statistics gives examples of bar charts without additional x-axis labels, Gustavii’s “How to Write and Illustrate a Scientific Paper†gives examples of bar graphs without them, Gary Klass’s web site (http://lilt.ilstu.edu/gmklass/pos138/datadisplay/sections/goodcharts.htm) , which seems to have consulted a lot of references, specifically recommends against an x-axis label in bar charts with labeled categories as completely unnecessary. But the kicker is probably that most (or at least a large fraction) of category bar graphs in Nature and Science do not have the redundant x-axis label. Nature and Science are not immune to bad graphs of course, but they’re pretty careful. So this criterion you’ve invented for a DAP is not however a criterion for a bad graph in the established view.
As for the unjustified assumption, in my view, it’s not present in the graph. My assumption on seeing this kind of plot is that the bars are arranged by height, nothing more. The ordering of the bars by height is recommended at the above web site, and in the above guide. Examples of this in Science and Nature are a little more scarce (though I found a few), mainly because in most cases the same categories are plotted against different variables, and it makes sense to keep the order of the categories the same. So the graph is a perfectly standard way to compare %DNA… for different categories. It is in the narrative where the assumption of a correlation of complexity from left to right is made, and whether or not that is justified does not make the graph misleading. So, your first criterion is bogus, and the second isn’t met.
“And in a later one:

”Dog’s Ass Plot (DAP, or Dapper):

A graphical representation of data in any field that, through a lack of clear axis labels, selective inclusion/exclusion of data, visual presentation style, and/or other questionable characteristics, generates a misleading interpretation of the data in the viewer, especially by implying an illusory pattern that is not supported by the available data.”â€
OK, sure, if a graph lacks clear labels, is selective of data, is questionable, misleading, and illusory, then it’s a bad graph. But when you criticized the graph in question, you didn’t convince me that it was any of these things. You complained about the overlap of the groups, but that doesn’t mean the groups can’t have a meaningful average, and you complained about the lack of labels and implied complexity, with which I disagreed in detail above.
Look at the “bar” for vertebrates. It slopes upward, culminating at the human value. These are not means or summary statistics like you see in bar graphs. This is implying an axis that is not labeled. It implies that humans are higher than all other vertebrates. And we don’t need to speculate about what the graph is meant to show, because it is given in the legend.
I don’t know why you’re hung up on the lack of axis labels as a single issue, because I did not establish that as a lone criterion. I said that if the figure, through lacking axes or other characteristics, implies a pattern that isn’t supported by the data, then it is a DAP.
You’re free to like this figure if you wish, but in my view it is nonsensical.
“Look at the “bar” for vertebrates. It slopes upward, culminating at the human value. These are not means or summary statistics like you see in bar graphs. This is implying an axis that is not labeled.â€
I admit the sloped bars suggest that some additional quantity is increasing from left to right, but I think that suggestion is there without the sloped bars. Any time you order the bars by height, you might expect there to be a reason for the order. If you can identify a quantity that increases left-to-right, then you have identified a correlation. You may quarrel with the identification, but I don’t see that as a problem with the graph.
“It implies that humans are higher than all other vertebrates.â€
Unless the data are wrong, humans *are* higher than all other vertebrates on the %junkDNA scale. The graph itself does not imply any other correlation.
Complexity may be difficult to quantify, but to this non-biologist, it seems obvious that the evolutionary scale of something increases left to right in that graph, and doesn’t everyone agree that humans are on top of the intelligence scale.
Of course, if the data are wrong, and I imagine it’s not easy to determine an average %DNA for all vertebrates, then it’s a bad graph by anyone’s criteria.
“And we don’t need to speculate about what the graph is meant to show, because it is given in the legend.â€
Yes, the authors are using the graph to demonstrate a correlation between complexity and y=%DNA…. They plotted y for various categories, in order of increasing y, and then claim the order corresponds to increasing complexity. This is completely standard procedure. They may have made a mistake in ranking complexity, but that’s a different problem.
“I said that if the figure, through lacking axes or other characteristics, implies a pattern that isn’t supported by the data, then it is a DAP.â€
And I’ve argued the graph does no such thing, even if the authors misinterpret the graph.
“You’re free to like this figure if you wish, but in my view it is nonsensical.”
I think you’ve let your objection to the claim that humans are the most complex influence your judgement on the quality of the graphical illustration.
Unless the data are wrong, humans *are* higher than all other vertebrates on the %junkDNA scale. The graph itself does not imply any other correlation.
Complexity may be difficult to quantify, but to this non-biologist, it seems obvious that the evolutionary scale of something increases left to right in that graph, and doesn’t everyone agree that humans are on top of the intelligence scale.
A DAP is a graph that, for whatever reason (including but not limited to lack of axes) implies a pattern that does not exist. Therefore, one must determine two things: 1) does it imply a pattern?, 2) is the pattern inaccurate?
You answered the first. Yes, it implies that there is a strong link between non-coding DNA amount and complexity. I will not disagree that humans are the most complex on intuitive measures. Do they have the most DNA? No, they don’t. Not by a long shot. So, the pattern strongly implied in the graph, which is further emphasized by having slopes on top of the bars to implies not means but ranges, is biologically nonsensical, and thus this is a DAP under the definition of the term (which, by the way, is not technical — it is tagged as “humour”).
“A DAP is a graph that, for whatever reason (including but not limited to lack of axes) implies a pattern that does not exist. Therefore, one must determine two things: 1) does it imply a pattern?, 2) is the pattern inaccurate?

You answered the first. Yes, it implies that there is a strong link between non-coding DNA amount and complexity.”
No, I didn’t say this, and I don’t believe the graph, by itself, implies it. The graph orders certain groups by %non-coding DNA (and I’m assuming this is right, or why bother with the graph at all). That’s it. The only way the graph can imply a link with complexity is if complexity increases with the order of the groups. You say it doesn’t, so the graph doesn’t imply a link.
Now, to someone who thinks, for whatever reason, that complexity increases with the order of the groups, then the graph together with this erroneous thought imply a link. For those who hold these incorrect ideas, the graph has been used effectively, as I see it. If, as you say, these ideas are wrong, and complexity doesn’t increase with the order of the groups, then it’s the idea that complexity increases with the order of those groups that should be criticized, not the graph itself. The intuition that I brought to the interpretation of the graph did not come from the graph. If you had listed those groups at random, and asked me to put them in order of intelligence, I’d have come up with the same order.
“I will not disagree that humans are the most complex on intuitive measures. Do they have the most DNA? No, they don’t. Not by a long shot.â€
But even if one assumes increasing complexity with the order of the groups, one doesn’t have to assume increasing complexity is linked to the amount of DNA.
I suppose criticism of the use of the graph is justified if it is exploiting a well-known but mistaken intuition to deceive non-experts. But my impression was that you were criticizing the presentation style of the graph, not the motivation of the graphers.
I admit the sloped bars suggest that some additional quantity is increasing from left to right, but I think that suggestion is there without the sloped bars. Any time you order the bars by height, you might expect there to be a reason for the order. If you can identify a quantity that increases left-to-right, then you have identified a correlation.
This graph implies a correlation between complexity and amount of noncoding DNA — and the legend confirms that this was the intention. This correlation does not exist, except when presented using selective data and with groups ordered misleadingly in this way. There are plenty of protists, invertebrates, plants, and other vertebrates with much larger genomes than humans. The figure does not present means, it shows sloping lines that imply ranges. It puts humans — the most complex — at the far right, but neglects to note that humans are NOT the highest when it comes to noncoding amount. Therefore it is a DAP, as I have defined the (whimsical) term.
“This graph implies a correlation between complexity and amount of noncoding DNA — and the legend confirms that this was the intention. “
The legend interprets the graph with the additional assumption that the ordering of the groups by bar height is consistent with increasing complexity. The graph does not mention complexity. The reader has to add that.
“This correlation does not exist, except when presented using selective data and with groups ordered misleadingly in this way. There are plenty of protists, invertebrates, plants, and other vertebrates with much larger genomes than humans. The figure does not present means, it shows sloping lines that imply ranges. It puts humans — the most complex — at the far right, but neglects to note that humans are NOT the highest when it comes to noncoding amount. Therefore it is a DAP, as I have defined the (whimsical) term.â€
OK, I can’t comment on the selectivity or correctness of the data, and if the graph is selective or incorrect, then obviously it’s a bad graph.
I started out disagreeing with *some* of your objections to the graph, namely, the absence of an x-axis label, and the overlapping of the categories, and the implied correlation with an unnamed quantity. I still disagree with these objections.
I don’t know how to say it more clearly: It is a DAP if it misleads the viewer to interpret a pattern that is not supported by the data. This can happen several ways, including lack of axis labels, strange categories, or selective datasets. None of these is, by itself, being criticized in general unless it contributes to an incorrect interpretation of data. This graph does, and it does so using several of those, so it is a DAP.
I repeat: a DAP is a figure that misleads by any of several possible factors, which by themselves may or may not be problematic in any particular situation. The specific trees are not important, I am talking about the forest. This figure is a DAP because it misleads the reader into thinking there is a correlation between DNA amount and complexity, and a lack of labels, sloped bars, giving humans separate from two other categories of which they are part, and omitting contradictory information all contribute to this explicitly intended interpretation.
My disagreement with your objections to particular aspects of the graph was not because those aspects are acceptable in isolation, but because I don’t think they make any negative contribution at all: In my opinion, the labels on that graph are perfectly clear, it doesn’t matter that the categories overlap, and it is useful to order the bars according to bar height. I didn’t mention the slanted bars at first, but I don’t have a problem with them either. If the categories have a range of y-values, that can be expressed nicely by slanted bars; since the bars are ordered by height from left to right, it makes sense to express the range with a slant increasing from left to right. If the species within a category were plotted separately, and ordered by bar height, you’d get the same result. Nothing in these aspects of the graph contributes to any implication of complexity. It’s not mentioned in the graph. If the data are selective, or incorrect, or if the graph takes advantage of well-known misconceptions, criticize it for those failings; no need to make up others.
Anon., why so obtuse? You must be bored to amuse yourself by annoying people with fake stupidity. It is fake, right?
Dear TR Gregory,
You seem to be confusing 'Genome Size' with 'Percent non-coding DNA'.
You seem to be confusing 'Genome Size' with 'Percent non-coding DNA'.
Not really. The two are more or less interchangeable, unless you assume a major difference in gene number and/or mean exon size. For vertebrates, these are likely to be very similar.
Polyploidy can also make percent non-coding DNA a poor proxy for genome size.
Perhaps you could comment on a similar figure produced by John Mattick and colleagues for BioEssays (29:288 – 299, 2007). This figure uses a separate column for each species. Does this address some of your concerns?
Recent polyploidy would add a second genome, but it would not change genome size. Ancient polyploidy, in which the genome behaves as a diploid (e.g., most gene duplicates inactive or divergent), would count as a change in genome size and would be less of a problem.
In any case, none of the organisms he depicts are likely to be (recent) polyploids, so it's a moot point. And all the other critiques of the depiction remain even if they were.
Perhaps you could comment on a similar figure produced by John Mattick and colleagues for BioEssays (29:288 – 299, 2007). This figure uses a separate column for each species. Does this address some of your concerns?
No, it does not. It's also a DAP because it has an unlabeled x-axis that tries to sneak an assumption in — namely that species at the right are a) most complex, and b) have the biggest genomes. It's also a very select sample that helps his case.
Fair enough.
Regarding the sample being select – accurate estimates of the amount of coding sequence are only available for a select set of species that have been sequenced and extensively annotated. Are there any such species that you think should have been included?
Given that genome size is a pretty reasonable stand-in for the percentage of non-coding DNA, you should be considering the full range of genome sizes. In that case, the pattern completely dissolves. So take your pick: add grasshoppers or onions or salamanders or algae or pufferfishes or hummingbirds whatever you like.